Publications
Work by our team and partners
Biotia team members have published over 40 peer-reviewed studies driven by expertise of sequencing technology spanning from clinical research on infectious diseases, to environmental metagenomics and microbial surveillance.
Featured
Publications
Publications
Thank you! Your submission has been received!
Oops! Something went wrong while submitting the form.
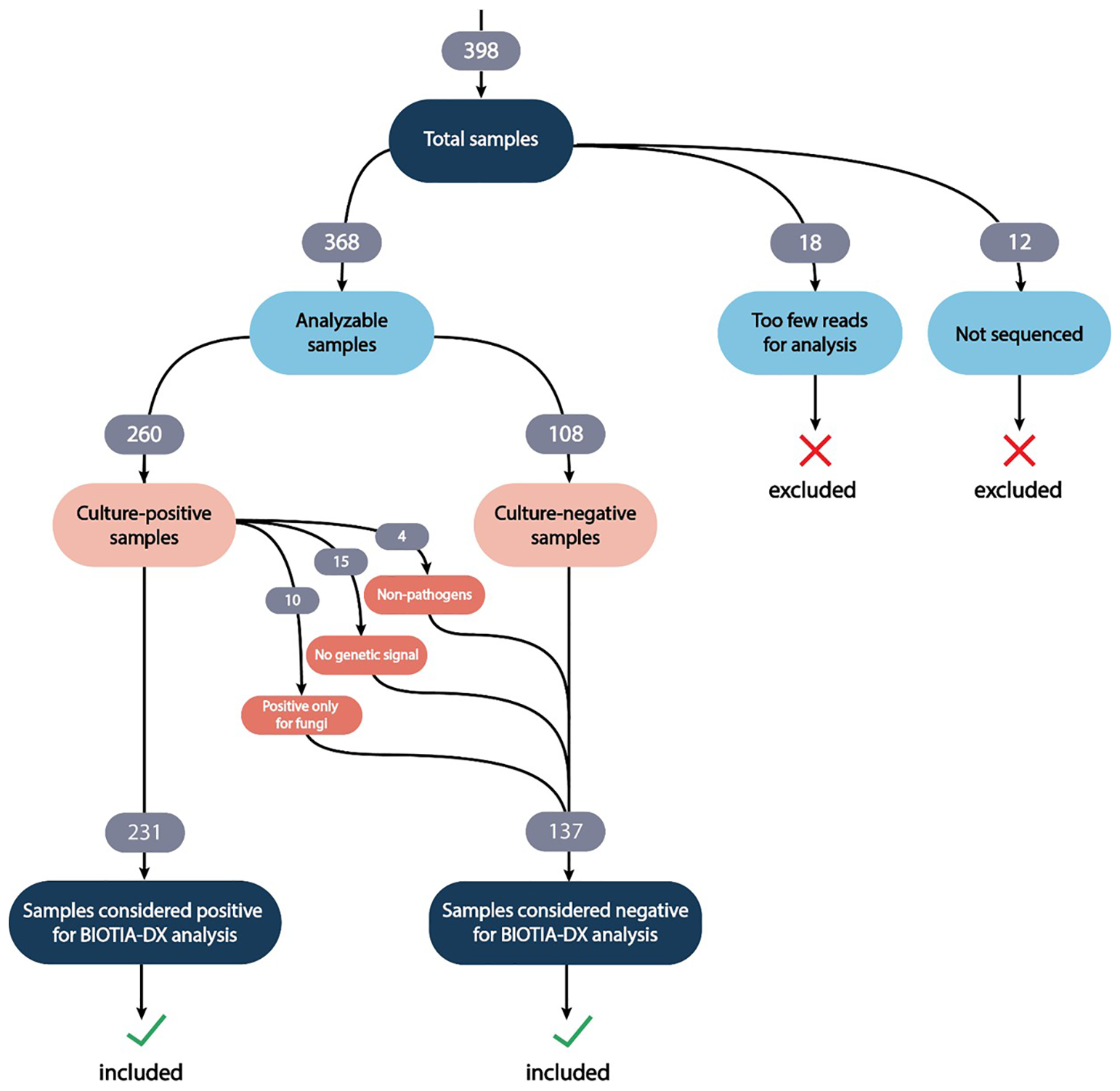
Analytical validation of a metagenomic next-generation diagnostic platform for urinary tract infection in a Thai tertiary hospital setting: a BI-Biotia UTI cohort study
The challenges and opportunities in creating a One Health warning system for pandemics

Potential Application of a Clinical-Grade Next Generation Sequencing Assay for Detection of Bacterial Vaginosis Associated Organisms in Urine

Comprehensive Next-Generation Sequencing-Based Assay for Antimicrobial Resistance Gene Marker Identification in Urine

ASM 2023: Highly Accurate and Reliable Next-Generation Sequencing-Based Urine Assay Overperforms Conventional Diagnostics

ASM 2023: A Global Pandemic Early Warning System

Utilization of Clinical-Grade Metagenomics in Urinary Tract Infection Diagnostics: Improving High-Risk Patients’ Outcomes with a Genomic-Based Assay
Investigation into the Potential Role of Cryptic Viral Infection in Villitis of Unknown Etiology Using Pan-Viral Metagenomic Sequencing

The Challenges and Opportunities in Creating an Early Warning System for Global Pandemics (pre-print)

Ozone Disinfection for Elimination of Bacteria and Degradation of SARS-CoV2 RNA for Medical Environments

ID Week 2022: Clinical-Grade Metagenomics in Urinary Tract Infections: Improving Performance of Next-Generation Sequencing Assays Using Internal Controls and Machine Learning

Comparative Effectiveness of Single vs Repeated Rapid SARS-CoV-2 Antigen Testing Among Asymptomatic Individuals in a Workplace Setting

Fecal Microbiota Transplantation Commonly Failed in Children with Co-Morbidities

IDWeek 2021: Performance Characteristics of Sequencing Assays for Identification of the the SARS-CoV-2 Viral Genome

IDWeek 2021: Extraction-free RT-PCR for detection of SARS-CoV-2 variants of concerns

IDWeek 2021: A novel likelihood-based model to estimate SARS-CoV-2 viral titer from next-generation sequencing (NGS) data

Draft Genome Sequences of Fungi Isolated from the International Space Station during the Microbial Tracking-2 Experiment

Sequencing of Circulating Microbial Cell-Free DNA Can Identify Pathogens in Periprosthetic Joint Infections

Characterization of Spacesuit Associated Microbial Communities and Their Implications for NASA Missions

Monoclonal Antibody Therapy in a Vaccine Breakthrough SARS-CoV-2 Hospitalized Delta (B. 1.617. 2) Variant Case

Draft Genome Sequences of Various Bacterial Phyla Isolated from the International Space Station

Evolution of Pathogen Response Genes Associated with Increased Disease Susceptibility During Adaptation to an Extreme Drought in a Brassica rapa Plant Population

Draft Genome Sequences of Aspergillus and Penicillium Species Isolated from the International Space Station and Crew Resupply Vehicle Capsule

Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions

Targeted Hybridization Capture of SARS-CoV-2 and Metagenomics Enables Genetic Variant Discovery and Nasal Microbiome Insights

Draft Genome Sequences of Lactobacillales Isolated from the International Space Station

IDWeek 2020 Abstract: SARS-CoV-2 NGS Assay Powered by Biotia COVID-DX Software

Draft Genome Sequences of Klebsiella Species Isolated from the International Space Station

End-To-End Protocol for The Detection of SARS-CoV-2 from Built Environments

Paenibacillus Infection With Frequent Viral Coinfection Contributes To Postinfectious Hydrocephalus In Ugandan Infants

Draft Genome Sequences of Enterobacteriales Strains Isolated from the International Space Station

Next Generation Sequencing Reveals the Skin Microbiome

Draft Genome Sequences of Rhodotorula mucilaginosa Strains Isolated from the International Space Station

Ozone Treatment For Elimination Of Bacteria And SARS-CoV-2 For Medical Environments (Preprint)

Global Genetic Cartography of Urban Metagenomes and Anti-microbial Resistance

A Metagenomic Analysis of Environmental and Clinical Samples Using a Secure Hybrid Cloud Solution (Abstract)

Bacteroides ovatus ATCC 8483 Monotherapy is Superior to Traditional Fecal Transplant and Multi-strain Bacteriotherapy in a Murine Colitis Model

Comparison of Five Testing Modalities for the Assessment of Patient Environment Cleanliness

Insights Into Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Phenotypes Through Comprehensive Metabolomics

Communicating the Promise, Risks, and Ethics of Large-scale, Open Space Microbiome and Metagenome Research (Preprint)

Metagenomic Characterization of Ambulances Across the USA

Comprehensive Benchmarking and Ensemble Approaches for Metagenomic Classifiers

Fecal Metagenomic Profiles in Subgroups of Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome

International Standards for Genomes, Transcriptomes, and Metagenomes

Serial Fecal Microbiota Transplantation Alters Mucosal Gene Expression in Pediatric Ulcerative Colitis

Composition and Function of the Pediatric Colonic Mucosal Microbiome in Untreated Patients with Ulcerative Colitis

The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium Inaugural Meeting Report

Transfer of Viral Communities Between Human Individuals During Fecal Microbiota Transplantation

Geospatial Resolution of Human and Bacterial Diversity with City-scale Metagenomics
